To Isolate Genomic DNA from Bacterial Cell
Theory:
- In molecular biology the isolation and purification of DNA from cell is most important process.
- The bacterial cell that is used in this procedure should be grown in suitable media under favourable conditions and harvested in late log to early stationary phase.
- Along with DNA bacterial cell contain RNA, lipids, protein which needs to separate.
- The first step in DNA isolation is to disrupt the cell membrane and this is done by SDS (Sodium Dodecyl Sulphate)
- Endogenous nucleases present on human fingertips can degrade the nucleic acid during purification. This degradation by nucleases can be prevented by chelating mg2+ ions using EDTA.
- Mg2+ ion is a necessary cofactor for the action of the nucleases.
- Proteinase K is used to degrade protein in the disrupted cell.
- And the function of phenol and chloroform is to denature and separate the protein from DNA.
- The denatured protein makes a layer between aqueous and the organic phase.
- DNA from the disrupted cell is precipitated by cold absolute ethanol or isopropanol.
Requirements:
- LB media
- E.coli DH5α cells
- TE buffer
- 10% SDS
- Proteinase K
- Phenol:Chloroform (1:1)
- 5M Sodium acetate
- Isopropanol
- 70%ethanol
- Autoclaved distilled water
- Eppendorf tube
- Micropipette
- Microtips
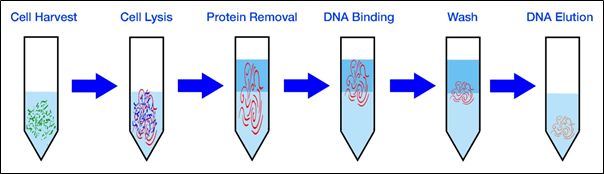
Procedure:
- Take 1.5ml of the bacterial culture (grown overnight) and harvest it by using centrifuge at 5000 rpm for 3-4 minutes.
- Discard the supernatant and add 600μl lysis buffer and incubate at room temperature for 10 minutes.
- Then add 200μl 10% SDS and 5μl proteinase K to the cells and incubate for 10-15 at 37o C.
- Now incubate the tubes in water bath for 10 minutes at 60o C and immediately transfer to ice bucket and keep for 5 minutes.
- Adding 500μl phenol: chloroform solution in 1:1 ratio and mix by inversion for 1-2 minutes, incubate for 5 minutes.
- After incubation centrifuge the mix at 10000 rpm for 10 minutes at 4o C.
- Now carefully transfer the upper aqueous layer to the fresh tube.
- Add 100μl 5M sodium acetate and equal amount of isopropanol mix by inversion and store at-20o C for 1 hour or overnight.
- Centrifuge the mix at 5000 rpm for 10 minutes and discard the supernatant.
- Then add 1 ml of 70% cold ethanol and mix by inversion.
- Again centrifuge at 5000 rpm for 10 minutes and discard supernatant.
- Air dry the pellet and add 40μl TE buffer and store at 4o C for further use.
Precaution:
- To avoid mechanical disruption of DNA cut tips should be used.
- The incubation period of proteinase K depends on the source of DNA and should be extended.

