INTRODUCTION:
- Mitosis is a type of cell division that takes place in living organisms and it is commonly defined as the process of duplication of chromosomes in eukaryotic cells and distributed during cell division.
- The process where a single cell divides resulting in two identical cells, each resulted cell contains the same number of chromosomes and same genetic composition similar to the parent cell.
- Mitosis was first discovered in plant cells by Strasburger in 1875. In 1879, mitosis is also discovered in animal cells by W. Flemming. Flemming in 1882 gave the term Mitosis.
- The term mitosis is derived from the Greek word such as ‘Mitos’ means thread.
OCCURRENCE OF MITOSIS:
- The mitosis takes place in somatic cells. The cells which undergo mitosis are called Mitocytes.
- In plants, the mitocytes are called meristematic cells. Some of the major sites of Mitosis in plants are root apex, shoot apex, intercalary meristem, lateral meristem, leaves, embryo, and seeds.
- In animals, the mitocytes are stem cells, germinal epithelium, and embryonic cells. In animals, it mainly takes place in Embryo, skin, and bone marrow.
- Mitosis also occurs during the regeneration of the cells. The mitosis takes place for three main reasons such as growth, repair, and asexual reproduction.
NUCLEUS:
It is a membrane-bound cell organelle present in both animal and plant cells. It is the center of the cell where genetic material is stored in the form of DNA. The DNA is arranged into a group of proteins into thin fibers. During the Interphase of the cell division, the fibers are uncoiled and dispersed into the chromatin. During mitosis, these chromatin condenses to become chromosome.
CHROMOSOME:
The chromosomes carry genetic material and they are made of DNA. The mitotic chromosomes possess two sister chromatids, they are narrow at the centromere. They also contain identical copies of original DNA. These Mitotic chromosomes are homologous, they are similar in shape, size, and location of centromere.

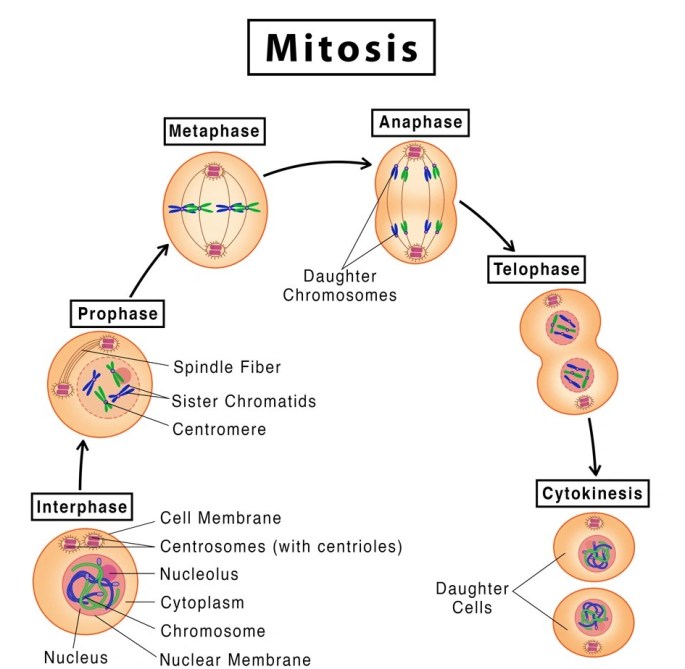
STAGES OF MITOSIS :
The mitosis cell division is broadly explained in two stages such
- Karyokinesis: Division of Nucleus. Greek ‘karyon’ means the nucleus, whereas ‘kinesis’ means movement.
- Cytokinesis: Division of Cytoplasm.
KARYOKINESIS:
4 different stages that take place in Karyokinesis.
1.Prophase
2.Metaphase
3.Anaphase
4.Telophase
PROPHASE:
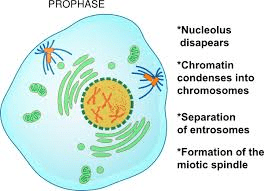
- The nucleus becomes spherical and the cytoplasm becomes more sticky.
- The chromatin slowly condenses into well-defined chromosomes.
- During Prophase, the chromosomes appear as a ball of wool. The chromosomes consists of two threads which are longitudinal known as chromatids.
- The chromosomes appear as two sister chromatids joined at the centromere.
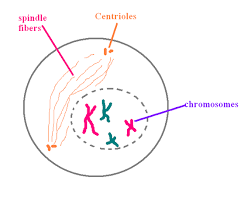
- The microtubules are formed outside the nucleus.
- In plant cells, the spindle apparatus is formed without centriole. In animal cells, the centriole is divided into two moves towards opposite poles.
METAPHASE:
- Nuclear envelop breaks down into membrane vesicles and the chromosomes are set free into the cytoplasm.
- Chromosomes are attached to spindle microtubules through kinetochore.
- Nucleolus disappears.
- Kinetochore microtubules arrange the chromosomes in one plane to form a central equatorial plate.
- Centromeres lie on the equatorial plane while the chromosome arms are directed away from the equator called auto orientation.
- Smaller chromosomes remain towards the center while larger ones arrange at the periphery.
- Metaphase is the longest stage of Mitosis and takes place for about 20 minutes. It is the best stage to study the structure of chromosomes.

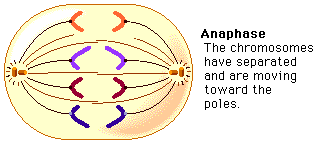
ANAPHASE:
- Chromosomes split simultaneously at the centromeres so that the sister chromatids separate.
- The separated sister chromatids move towards the opposite poles.
- The daughter chromosomes appear in different shapes such as V-shaped(metacentric), L-shaped(sub-metacentric), J-shaped(acrocentric), rod-shaped(telocentric).
- The spindle fibers are attached to the centromere and pull the chromosomes to the poles.
- Anaphase is the shortest stage of Mitosis.
TELOPHASE:
- Daughter chromosomes arrive at the poles. Kinetochore microtubules disappear.
- Chromosomes uncoil into chromatin.
- Nucleolus reappears. The formation of nuclear envelope occurs around each pair of chromosomes.
- The Viscous nature of the cytoplasm decreases.
- Telophase is called a reverse stage of the prophase.

CYTOKINESIS:
Cytokinesis is defined as the division of cytoplasm.
- It starts during the anaphase and is completed by the end of the telophase.
- It takes place in 2 different methods.
a) Cell plate method: It takes place in plant cells. The vesicles of Golgi fuse at the center to form a barrel-shaped phragmoplast. The contents of the phragmoplast solidify to become a cell plate, this cell plate separates the two daughter cells.
b) Cleavage or cell furrowing method: It takes place in Animal cells. In this method, a Cleavage furrow appears in the middle, which gradually deepens and breaks the parent cell into two daughter cells.


SIGNIFICANCE OF MITOSIS:
- Mitosis is called as an equational division in which daughter cells produced are identical.
- It maintains the constant number of chromosomes and genetic stability in somatic and vegetative cells of the living organisms.
- It helps to increase the cell number so that zygote transforms into a multicellular adult.
- Healing wounds takes place by Mitosis.
- It helps in asexual reproduction.
- Mitosis is necessary for growth, maturity and to repair damaged cells.
BY: ABHISHEKA (MSIWM013)