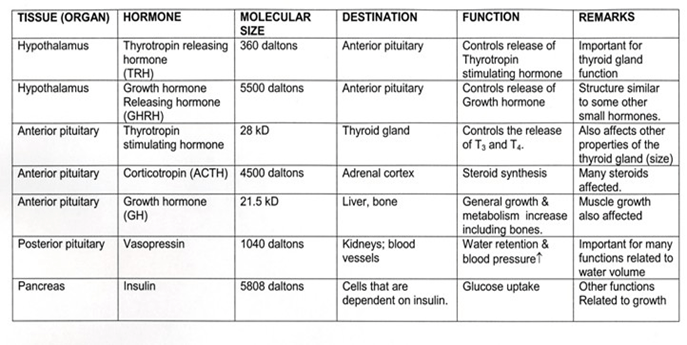
INTRODUCTION:
The removal of amino group from the amino acid as ammonia (NH3) is called deamination.
A chemical reaction that is catalysed by the deaminase class of enzymes which results in the liberation of ammonia is called deamination and this liberated ammonia is used for urea synthesis.
These reactions occur in the liver and kidney of humans. In the kidney, the ammonia which is a result of the conversion of the amine group (that is removed) is excreted from the human body.
Deamination can be either oxidative or non-oxidative.
OXIDATIVE DEAMINATION:
When an amine group is removed from a molecule by the process of oxidation, the reaction is called an oxidative deamination reaction.
These types of reactions largely occur in liver and kidney.
These reactions lead to the production of alpha-keto acids etc. via the amine groups.
This reaction is very important in the catabolism of amino acids as it forms a catabolized product from amino acids.
The by-product of this reaction is ammonia which is a toxic. Here, the amine group converts into ammonia. The ammonia that is formed is the transformed in urea which is an excretion product of the body.

The primary reactants of such a reaction are glutamic acid or glutamate. This is because usually the end product of most transamination reactions is glutamic acid.
Glutamate dehydrogenase is the enzyme involved that catalyzes the transfer of an amino group to an alpha-keto acid group.
Another enzyme involved in these reactions is the monoamine oxidase enzyme that catalyzes the deamination via the addition of oxygen.
- Glutamate dehydrogenase (GDH):
Glutamate dehydrogenase is a mitochondrial enzyme. It also contains the element zinc.
It contains six identical units and has a molecular eight of 56,000 each.
GDH is controlled by allosteric regulation where GTP, ATP, steroid and thyroid hormones inhibit GDH whereas GDP and ADP activate it.
- Glutamate dehydrogenase and its roles in the process of oxidative deamination:
Glutamate serves as a “collection centre” for amino groups in biological systems because the amino groups of most amino acids are transferred to glutamate.
Rapid oxidative deamination of glutamate leads to the production of ammonia. This is catalysed by glutamate dehydrogenase.
The importance if this GDH catalysed reaction lies within the reversibility of linking up glutamate metabolism with the tricarboxylic acid cycle. This reversibility is credited to the enzyme of alpha-ketoglutarate that is involved.
GDH is unique because it can utilize either NAD+ or NADP+ as a coenzyme.
The intermediate that is formed during the conversion of glutamate to alpha-ketoglutarate is iminoglutarate.
- Regulation of GDH activity:
The glutamate levels are increased in the body after the consumption of a protein-rich meal and glutamate is converted to alpha-ketoglutarate with the liberation of ammonia.
Further, the degradation of glutamate is increased when the cellular energy levels are low to provide alpha-ketoglutarate which enters the TCA cycle to liberate energy.
- Process of oxidative deamination by amino acid oxidases:
Alpha-keto acids and ammonia are produced by L-amino acid oxidase and D-amino acid oxidase flavoproteins which possess FMN and FAD respectively.
The result of this reaction is a reduced form of oxygen, which is H2O2.
This H2O2 then undergoes a decomposition reaction for which the enzyme is catalase.
L-amino acid oxidase does not act on glycine and dicarboxylic acids and therefore the activity of L-amino acid oxidases is much lower than that of D-amino acid oxidases.
- Fate of D-amino acids:
D-amino acids are found in plants and microorganisms but are not found in mammalian cells.
However, they are taken regularly in the diet and metabolised in the body by D-amino acid oxidases to produce respective alpha-keto acids by oxidative deamination.
The first step for the conversion of unnatural D-amino acids to L-amino acids is catalysed by D-amino acid oxidase and is therefore of value within the body.
NON-OXIDATIVE DEAMINATION:
The process of removal of amine groups from a molecule via reactions, all except the oxidation reactions, is called a non-oxidative deamination reaction.
The main types of reactions that are involved in this process are:
1. Reduction
2. Hydrolysis
3. Intramolecular reactions.
However, this reaction also involves the production of toxic by-product ammonia from amino acids.
Moreover, the most common amino acids that undergo this type of reactions are hydroxy amino acids (serine, threonine, cysteine and histidine), sulphur amino acids (cysteine and homocysteine). Similarly, the most common enzymes involved in this reaction are dehydratases, desulphahydrases, lyase, histidase etc.

The examples of non- oxidative deamination are:
(a) Amino acid hydrases: The hydroxy amino acids undergo deamination by PLP-dependent dehydrases to produce respective alpha-keto acids with the release of ammonia.
(b) Amino acid desulphhydrases: Form keto acids by undergoing a coupled reaction of sulfur amino acids undergoing deamination along with desulphhydration.
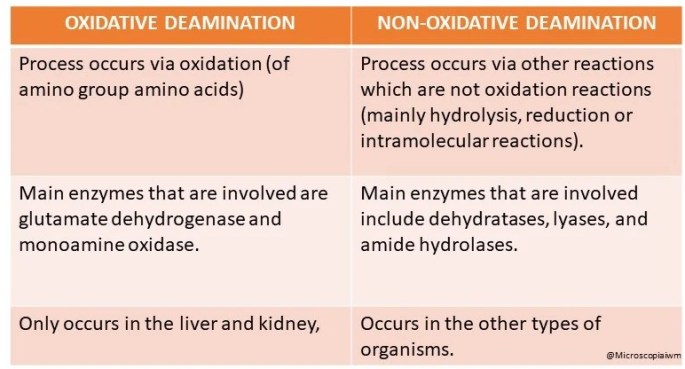
DIFFERENCE BETWEEN OXIDATIVE AND NON-OXIDATIVE DEAMINATION:
Difference in process:
1. Oxidative deamination: Process occurs via oxidation (of amino group amino acids).
2. Non-oxidative deamination: Process occurs via other reactions which are not oxidation reactions (mainly hydrolysis, reduction or intramolecular reactions).
Differences in the enzymes involved:
1. Oxidative deamination: Main enzymes that are involved are glutamate dehydrogenase and monoamine oxidase.
2. Non- oxidative deamination: Main enzymes that are involved include dehydratases, lyases, and amide hydrolases.

BY- Shaily Sharma (MSIWM041)