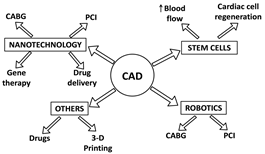
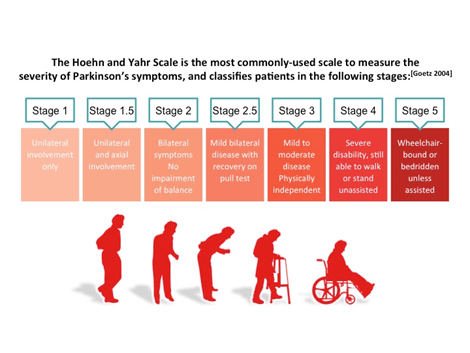
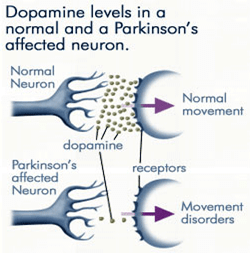
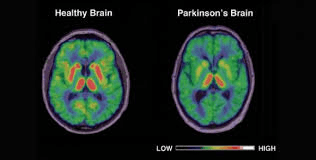
BY: K. Sai Manogna (MSIWM014)
In 1952, Joshua Lederberg invented the word ‘plasmid.’ It was initially formed from bacteria; plasmids are extrachromosomal genetic elements that can reproduce independently in most Archae, Eukarya species, and Eubacteria. Plasmids are double-stranded circular DNA molecules that are different from the chromosomal DNA of the cells.
The genetic material found within the chromosomal DNA guides the structure and function of a bacterial cell. In some instances, plasmids are usually not required for the host bacterium to survive. Although not necessary, by encoding functions that may not be defined by the bacterial chromosomal DNA, plasmids significantly contribute to bacterial genetic diversity and plasticity. Antibiotic tolerance and the expression of proteins, for example. The plasmid is also encoded by antibiotic resistance genes, allowing the bacteria to survive in an environment containing antibiotics, thereby providing the bacterium with a competitive advantage over species susceptible to antibiotics. Plasmids may be altered as a means to express the protein of interest for e.g., production of human insulin using recombinant DNA technology.
As a mechanism for gene-cloning and as a vehicle for gene-expression, plasmids have been central to modern recombinant DNA technology.
Isolation of Plasmids
In molecular biology, bacterial plasmid DNA isolation is a crucial technique and an integral step in many processes, such as cloning, DNA sequencing, transfection, and gene therapy. These manipulations need the isolation of high purity plasmid DNA. In all molecular biology procedures, cloning, such as digestion with restriction enzymes, PCR, transfection, in vitro translation, blotting, and sequencing, purified plasmid DNA can be used for immediate usage.
In molecular biology, alkaline lysis is used to separate plasmid DNA or other cell components, such as proteins, by splitting open the cells. Primarly, bacteria containing the plasmid of interest are allowed to grown and then lysed with an alkaline lysis buffer composed of a sodium dodecyl sulphate (SDS) detergent. Cellular debris is extracted, and the plasmid is separated and filtered through a series of steps, including agitation, precipitation, centrifugation, and the removal of the supernatant.
Principle:
Purification of plasmid DNA from bacterial DNA using alkaline lysis is based on the differential denaturation of plasmid and chromosomal DNA. Disruption of the cellular structure to produce a lysate, separation of the plasmid from the chromosomal DNA, cell debris, and other insoluble material are the essential steps of plasmid isolation. With a lysis buffer solution, bacteria are lysed.
Materials and Equipment:
Refrigerated centrifuge
Vortex
Microwave oven
pH meter
Orbital shaker
Micropipettes
Autoclave
LB plate with Bacterial colonies
1.5 ml micro-centrifuge tubes
Autoclaved distilled water
Microtips
Microfuge tubes
Chemicals:
1. Lysis Solution (Solution I)
2. Denaturation solution (Solution II)
3. Neutralizing solution (Solution III)
4. TE Buffer
5. RNase
6. Phenol: Chloroform: isoamyl alcohol
7. 70% Ethanol
8. Isopropanol
Preparation of Stock solutions:
- Solution I (Lysis solution): 50mM Glucose, 25mM Tris-Hcl (pH 8.0), 10mM EDTA (pH 8.0). Store at 4oC.
- Solution II (Denaturation solution):1% SDS, 0.2N NaOH (pH 12.0). Freshly prepared and store at room temperature.
- Solution III (Neutralizing solution): 60 ml of 5 M potassium acetate, 11.5 ml of glacial acetic acid, and 28.5 ml of water. Store at 4oC.
- TE Buffer: 10mM Tris-HCl (pH 8.0), 10mM EDTA (pH 8.0)
- RNase (1mg/ml)
- Phenol: Chloroform: Isoamyl alcohol -(25:24:1).
- Washing buffer
- Elution buffer
Biological material:
Overnight grown culture of E.coli.
PROCEDURE
Harvesting of the cells:
- The single bacterial colony is transferred into a 5 ml LB medium containing appropriate antibiotic, incubated for 16 hrs at 37oC with vigorous shaking.
- About 1.5 ml culture is transferred into a microfuge tube and centrifuged at 6000 rpm for 5 min at 4oC and discard the supernatant.
Isolation of Plasmid by alkyl-lysis method:
1. The bacterial pellet is re-suspended in 100μl of ice-cold solution-I by vigorous vortexing. This is essential to ensure that the bacterial pellet is wholly dispersed in the solution I.
2. Add 200μl of freshly prepared solution II. Mix the contents by inverting the tubes rapidly 4 to 5 times. A transparent and viscous solution ensured complete lysis. Do not vortex. Incubate on ice for 3 min.
3. Add 150μl of ice-cold solution III. Mix by gently inversion for about 10 seconds to disperse the solution III through the vicious bacterial lysate. Further incubate the tubes on ice for 5 min.
4. Centrifuge at10,000 rpm for 5 minutes at 4oC and transfer the supernatant to a fresh tube.
5. To this add equal volumes of phenol-chloroform, mix by vortexing. After centrifuging at 10,000 rpm 5 minutes at 4oC, transfer the aqueous phase to a fresh tube.
6. Precipitate the double-stranded DNA by adding an equal volume of isopropanol, mix gently, and allow the mixture to stand for two minutes. Then centrifuge at 10,000 rpm for 10 minutes at 4oC.
7. Remove the supernatant by gentle aspiration.
8. Then wash the pellet with 1 mL of 70% ethanol (twice). Allow the DNA pellet to air dry for 10 minutes.
9. Re-dissolve the DNA pellet in 50μl of TE (pH 8.0).
Isolation of PLASMID by spin column method:
1. Inoculate a single colony of bacterial cells from the petri plate using a sterile inoculation loop to 5ML of LB broth and allow them to grow overnight at 37°C at 200 rpm.
2. From the pre-inoculum transfer, 1.5 ML of bacteria-containing cells into 2mL centrifuge tubes and centrifuge the cells at 8000 rpm. for one minute. Then the supernatant was discarded.
3. Re-suspend the cells in 200 µL of resuspension solution. Then mix the cells thoroughly by pipetting up and down or vortexing it.
4. To this, add 200 µL of lysis solution and mix the tubes by gently inverting up and down.do not vortex it. They were then incubated for five minutes.
5. To this, add 350 µL of the neutralizing solution and, for mixing, invert the tubes 4 to 6 times. Then Centrifuge these tubes at 10,000 rpm for 10 minutes.
6. Binding column preparation: Take a new centrifuge tube and insert a new binding or spin column into it. To this, add 500 µL of column preparation solution and spin at 10,000 rpm for one minute. Then discard the solution that was collected in the centrifuge tube.
7. Then, transfer the clear lysate to the binding column and centrifuge at 10,000 rpm for one minute. Discard the flow-through that was remained in the centrifuge tube.
8. Washing: In this step, add 750 µL of washing solution to the binding column and spin the centrifuge tube for one minute. Then discard the solution that remained in the centrifuge tube. (Do the washing step twice).
9. Then, change the spin column into the new centrifuge tube. Then spin the column for one minute and incubate at room temperature for 2 – 5 minutes.
10. ELUTION: In this step, the spin column is transferred to the fresh collecting tube. Then add 70 µL of elution buffer to the spin column and spin for one minute at 10,000 rpm. Label the vial as elution I.
11. Transfer the same spin column to the fresh collecting tube and add 40 µL of elution buffer to the spin column and spin for one minute at 10,000 rpm. Label the vial as elution II.
12. Then discard the spin column, and the vials labelled as elution I and II are stored at -20oC or -80oC.

Preparation of agarose gel electrophoresis:
- First, take a gel casting tray, then clean the tray with ethanol and seal the edges with transparent tape and place the comb.
- Then 0.8% of agarose solution was prepared using 1X TBE. When the temperature was lowered, add a little amount of Ethidium bromide to it and mix well.
- After mixing, pour the agarose solution into the gel casting trays and then allow for polymerization. Make sure with the absence of air bubbles while pouring the solution. After polymerization, remove the comb without breaking the gel.
- Submerge the agarose gel into the electrophoresis tank containing 1X TBE buffer.
Electrophoresis of isolated plasmid DNA.
1. Take 3 – 4 µL of eluted sample and add 2 µL of sample buffer.
2. Load 5-6 µL of a sample into each well along with a DNA marker or ladder.
3. Run the electrophoresis and allow the plasmid DNA to run in 1X TBE buffer at a constant voltage. Keep track of the dye front.
4. Then disconnect the power electrophoresis tank and remove the gel using gloves. Place the gel in the Gel Documentation System or Transilluminator and visualize the DNA bands in the UV light.
5. Quantify the concentration of DNA from the DNA marker or ladder used.

Role of chemicals used:
Spin column: The desired nucleic acids should be bound to the column after centrifuging the lysate through the silica membrane, and impurities such as protein and polysaccharides should be in the flow-through. While plant samples are likely to contain polysaccharides and pigments, the membrane may be slightly brown or yellow in blood samples. The washing steps will remove such impurities. Usually, there are two washing measures, but this varies depending on the type of sample. A low concentration of chaotropic salts to eliminate residual proteins and pigments will also be part of the first wash. In order to extract the salts, this is often accompanied by an ethanol wash. Columns contain a resin of silica which binds to DNA/RNA selectively. By its capability to bind silica in the presence of high concentrations of chaotropic salts, the DNA of interest can be isolated.

Washing: With an alcohol-based wash, these salts are then removed, and the DNA is eluted using a low-ionic-strength solution such as TE buffer or water. Dehydration and the formation of hydrogen bonds that compete against poor electrostatic repulsion seem to be driving the binding of DNA to silica. Therefore, a high salt concentration will help push DNA adsorption to silica, and the DNA will be released at a low concentration.
Elution buffer: The Elution buffer is initially used to wash away unbound proteins and release the ligand’s desired protein at a higher concentration. The elution buffer must work rapidly without altering the desired protein’s function or activity. This enables to stand within the membrane for a few minutes before centrifugation for optimum DNA elution.
Glucose: It helps in maintaining osmolarity and keeps the cells from bursting.
Tris HCL and EDTA: This helps in chelating divalent metal ions such as magnesium calcium and destabilizes the cell wall, inhibiting DNases’ action.
NaOH and SDS: In the lysis buffer or solution II, sodium hydroxide, and the detergent Sodium Dodecyl (lauryl) Sulfate (SDS) are present. SDS is used to solubilize the cell membrane. NaOH helps in the breakdown of the cell wall, but more significantly, it interferes with the hydrogen bonding between the DNA bases, turning the cell’s double-stranded DNA, including the genomic DNA and the plasmid, into single stranded DNA.
RNase: It helps incomplete digestion of unwanted RNA from the plasmid sample.